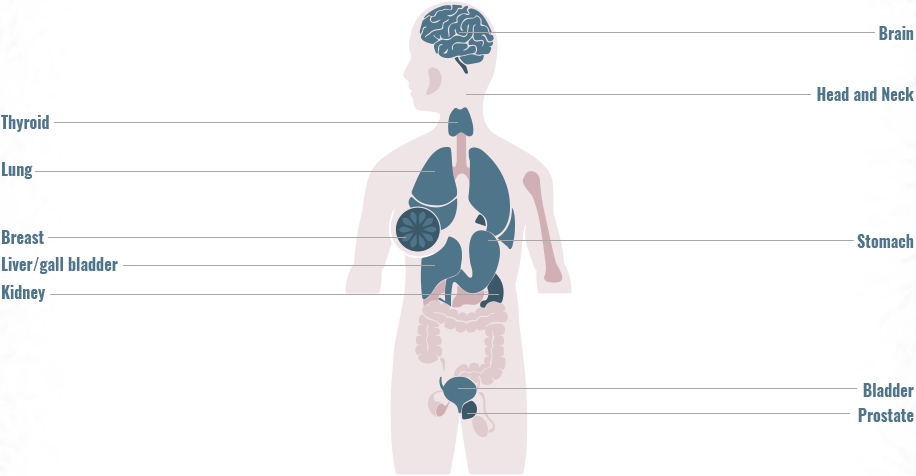
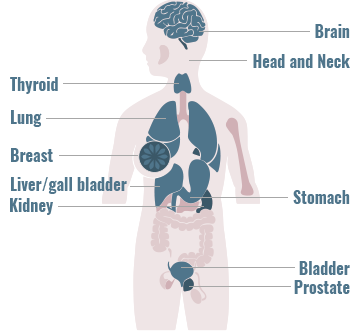
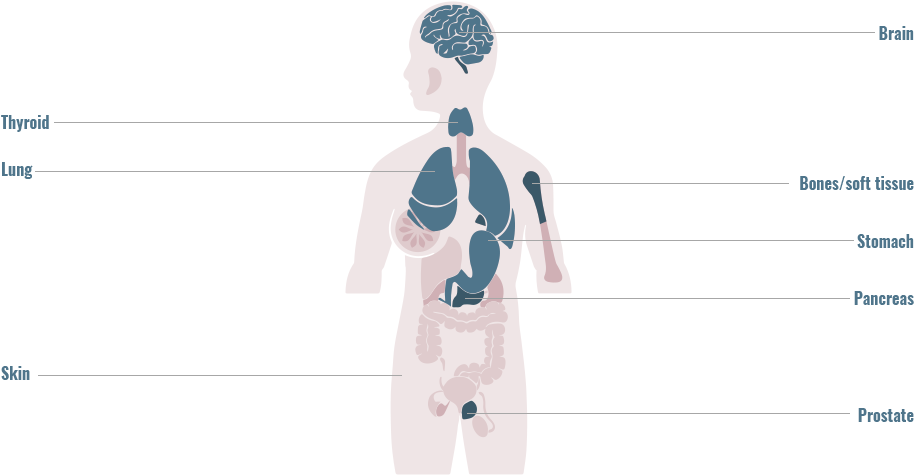
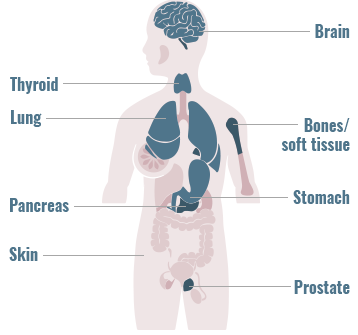
Precision Medicine has particular relevance in oncology
Precision Medicine in oncology is the use of therapeutic interventions to identify and treat patients based on specific molecular or cellular features of their cancer, such as genomic alterations or protein expression levels.1
Advancements in testing have led to the identification of multiple genomic alterations, changing the approach to cancer treatment2-5
Rapidly advancing research and technology, especially the sequencing of human and cancer genomes, has identified numerous potential genomic and biologic targets that are moving patient care toward Precision Medicine.6
- Studies suggest that 30% to 49% of patients with cancer who undergo genomic profiling have an actionable alteration, defined as having potential to be paired with an approved or investigational therapy7,8
-
Most actionable genomic alterations:
- Are uncommon5,9
- Occur in a mutually exclusive manner10
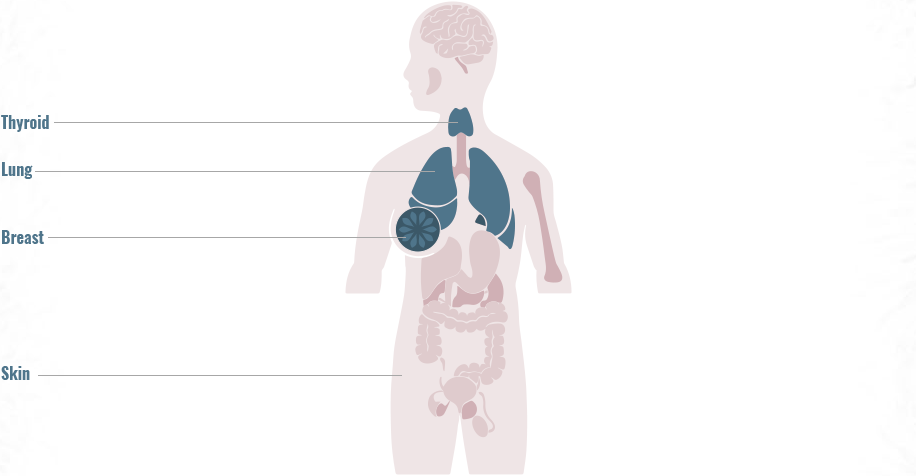
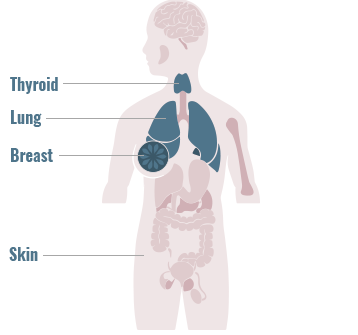
Genomic alterations are often uncommon, highlighting the need for comprehensive genomic testing to match patients to approved or investigational therapies5,9
Test Your Knowledge

Select all that apply.
NTRK, neurotrophic tyrosine receptor kinase.
References: 1. Yates LR, Seoane J, Le Tourneau C, et al. The European Society for Medical Oncology (ESMO) Precision Medicine Glossary. Ann Oncol. 2018;29(1):30-35. 2. Schram AM, Chang MT, Jonsson P, Drilon A. Nat Rev Clin Oncol. 2017;14(12):735-748. 3. Ross JS, Wang K, Gay L, et al. Oncologist. 2014;19(3):235-242. 4. Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media, LLC; 2014:3-11. 5. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. 6. Kumar-Sinha C, Chinnaiyan AM. Nat Biotechnol. 2018;36(1):46-60. 7. Boland GM, Piha-Paul SA, Subbiah V, et al. Clinical next generation sequencing to identify actionable aberrations in a phase I program. Oncotarget. 2015;6(24):20099-20110. 8. Massard C, Michiels S, Ferté C, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586-595. 9. Kummar S, Lassen UN. Target Oncol. 2018;13(5):545-556. 10. Cisowski J, Bergo MO. What makes oncogenes mutually exclusive? Small GTPases. 2017;8(3):187-192.
Some active gene fusions are identified as actionable alterations in cancer1
One important type of genomic alteration is gene fusion, which like other alterations can lead to oncogene addiction, in which cancers are driven primarily, or even exclusively, by aberrant oncogene signaling.1
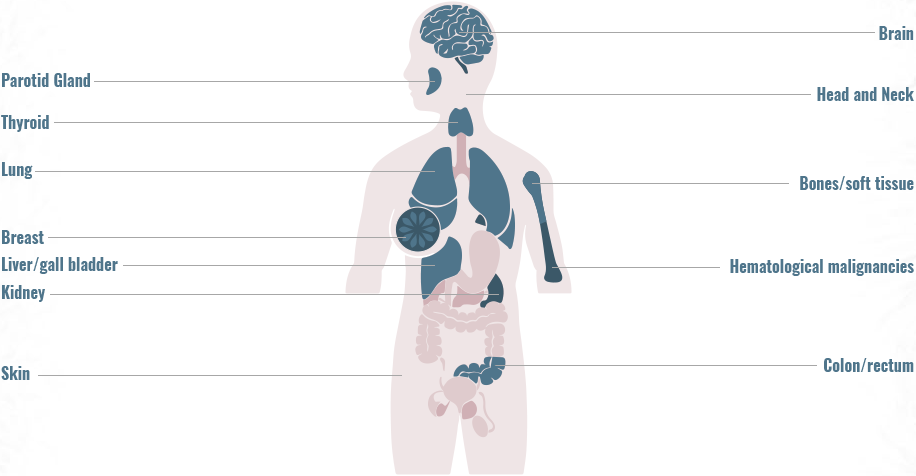
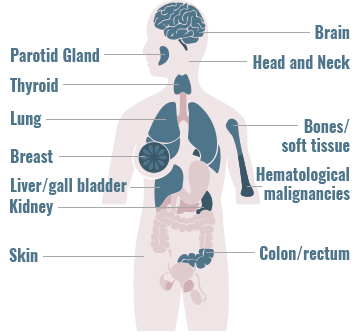
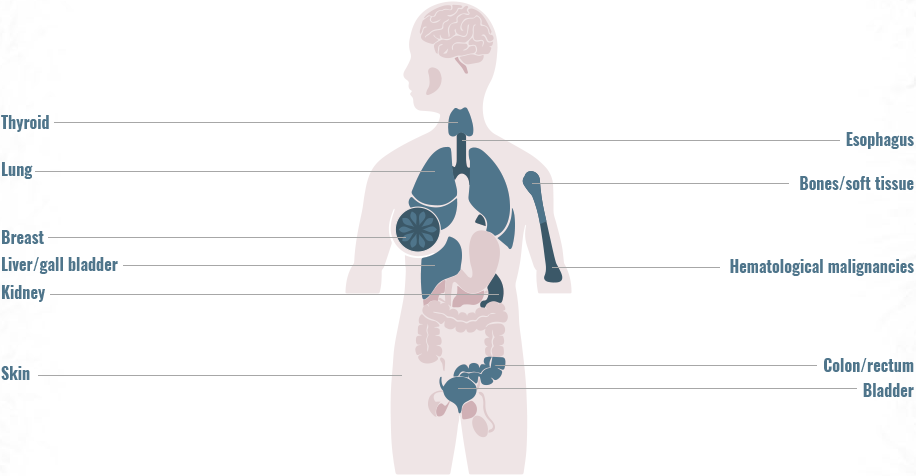
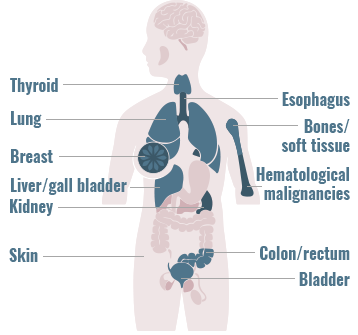
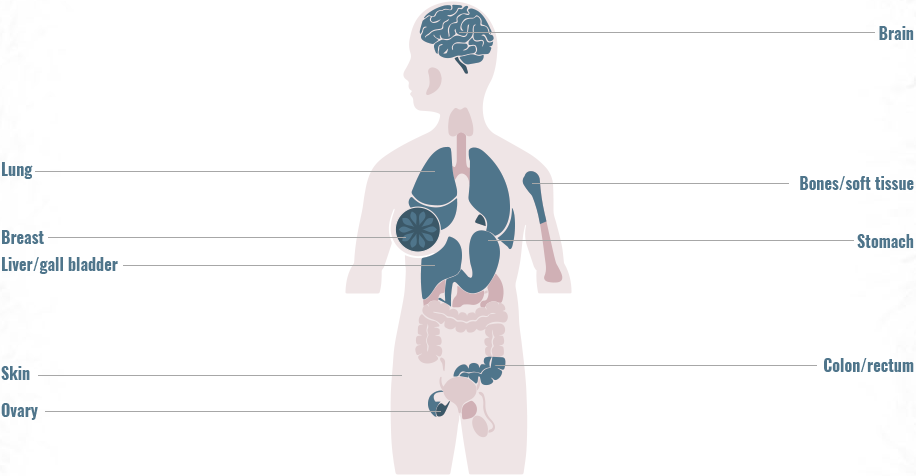
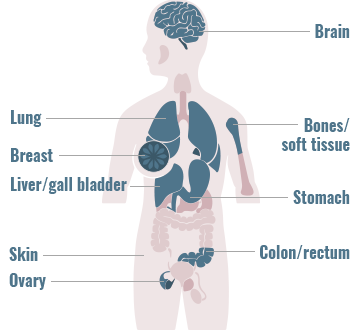
Gene fusions can be found across a diverse range of solid and hematologic malignancies1-4
- Gene fusions account for approximately 20% of cancer morbidity2
- ~10,000 unique gene fusions have been identified (only a small number resulting in actionable driver oncogenes)2
- >90% of these were identified during the past few years due to advances in deep-sequencing and fusion detection algorithms2
-
The frequency of gene fusions varies by gene fusion and tumor type
- 90% of lymphomas harbor gene fusions2
- Up to 10% of NSCLC tumors harbor gene fusions5
Discovery of actionable gene fusions opens doors for patients to better understand the next steps in their cancer care2
Test Your Knowledge

Select all that apply.
ALK, anaplastic lymphoma kinase; BRAF, proto-oncogene B-Raf; CRAF, proto-oncogene C-Raf; FGFR, fibroblast growth factor receptor; NSCLC, non-small cell lung cancer; NTRK, neurotrophic tyrosine receptor kinase; RET, Ret proto-oncogene; ROS1; ROS proto-oncogene 1.
References: 1. Schram AM, Chang MT, Jonsson P, Drilon A. Nat Rev Clin Oncol. 2017;14(12):735-748. 2. Latysheva NS, Babu MM. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016;44(10):4487-4503. 3. Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58-66. 4. Ross JS, Ali SM, Fasan O, et al. ALK fusions in a wide variety of tumor types respond to anti-ALK targeted therapy. Oncologist. 2017;22(12): 1444-1450. 5. Farago AF, Taylor MS, Doebele RC, et al. Clinicopathologic features of non–small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol. 2018.